Jump To:
Introduction
Closed-Source Genetics
Open-Source Genetics
Going Public With My Genome
Better Living Through Personal Genomics
DIY Genomic Sequencing for Programmers
My Personal Genomic Results
Further Reading
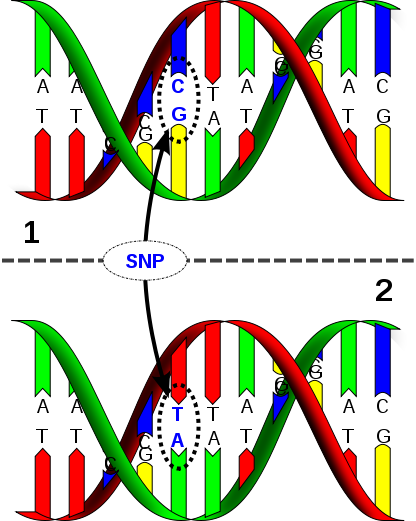
Single Nucleotide Polymorphism (SNP)
Introduction
It’s been over a year since I signed up with 23andMe and several months now since I downloaded my raw genomic data from them and started seeing what I could learn from it on my own. Although very few services out there will fully sequence your personal genome, by focusing on sequencing an individual’s Single Nucleotide Polymorphisms (SNPs), pronounced “Snips” for short, which are the variables between human genomes, we can focus on what’s of interest in our personal genomic data and get this data relatively cheaply. By comparing the differences in our genes, our genotypes, my wife and I could learn about how they differently express themselves in our lives, our phenotypes, to gain insights about our health risks as well as interesting traits about ourselves that may explain our behaviors and experiences throughout life.
Closed-Source Genetics
For $1,100 deCODEme will “Calculate your genetic risk for 47 conditions and traits,” but not include any ancestry analysis. Alternatively, Navigenics offers genome scans for $2,750 according to a 2009 Wired article (they don’t post prices online themselves), and Knome offers total genome sequencing for $3,750.
So at $207 test + year-subscription deal 23andMe has far and away the best deal for getting your personal genome sequenced. Although 23andMe doesn’t fully sequence your genome, it does sequence 960,520 SNPs and you get to download the whole thing to keep and take with you–which is crucially important (as I will get to in the open-source part further down).
The Good Stuff
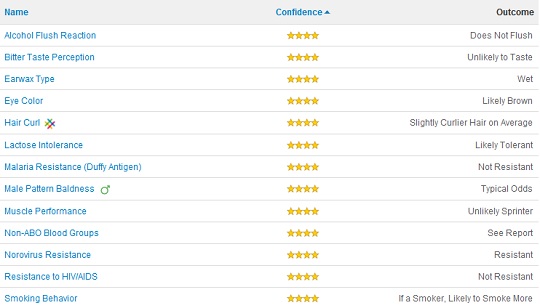
My Traits
The best thing about 23andMe is the interface. They take your results and break them down into your disease risks, drug responses, and various traits. Right off the bat it impresses me for correctly identifying my phenotype as expressing brown eyes (rs12913832(A;G)), curly hair (what hasn’t fallen out (rs17646946(G;G))), lactose tolerance (rs4988235(A;G)), and a tendency to smoke like a chimney if I smoked (which I did at a pack or more a day for 10 years (rs1051730(A;G))).
But it’s the stuff you don’t know about yourself that makes it worth it. I learned that I cannot perceive bitter tastes (rs713598(C,C)), which was interesting, while my wife’s apparent ability to taste bitter could explain some of our differing culinary preferences (although we couldn’t think of anything). Then there was the norovirus resistance (rs601338(A;A)), which could explain why I usually get off easy when a stomach flu goes around.
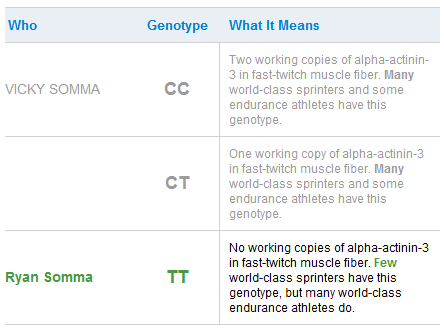
ACTN3
But most enlightening was finding I lack any copies of alpha-actinin-2 (ACTN3) in my fast-twitch muscle fiber (rs1815739(T;T)), meaning I am a terrible sprinter and throughout my public school years I was indeed consistently a poor-performer at short-distance sprints. As my father, a runner of marathons now approaching his 70s, liked to say, “In the running world there are gazelles and there are plough horses. I am a plough horse.” Now I know that my inability to make a half-marathon in less than two hours is likely hereditary. At the same time, the TT genotype may confer some ability to make greater gains at strength training, which would explain my enjoyment of weight lifting.
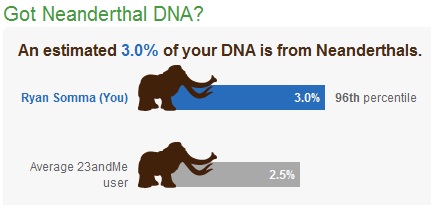
Neanderthal DNA
23andMe also offers some fun stuff, like calculating how much Neanderthal DNA you have. I was proud to learn that I share 3.0% of my genes with our distant cousins, putting me in the 96th percentile, which I hope means that my ancestors made a bigger contribution to breeding them out of existence.
The Neutral Stuff
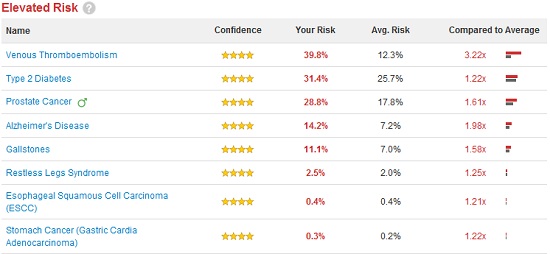
Disease Risk
According to the “Disease Risk” section of 23andMe, I’m 1.22 times more likely than the average person to develop Type II Diabetes. If this was Type I Diabetes, which is purely genetic, this result would mean something to me, but Type II is largely a matter of lifestyle. So what does it mean for me to have an elevated risk? The site says that the disease is only 26% heritable, meaning if I eat right and exercise, I should be fine… right? According to this survey my risk is extremely low and my BMI is borderline–but then BMI is useless for people who lift weights. Then there’s also the fact that, drilling down into the data, I find this “risk” is computed using 11 genetic markers, the red ones show increased risk, the green ones decreased risk:
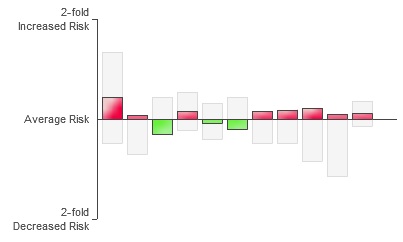
Diabetes Markers
I’m ambivalent about the usefulness of this data. It’s really difficult to determine how concerned or unconcerned I should be or even how seriously I should take this information. Apparently I’m also at risk for a greater tendency to overeat on average if I were an “Old Order Amish woman.”
But these are less criticisms of 23andMe and more problems with the whole of personal genomics in general. The research is still coming in and will continue coming in for decades, maybe even centuries. So it’s important to not just accept the initial third-party extrapolations from your genetic data, but to check in on it from time to time and learn what new developments have come up.
The Bad Stuff
23andMe Parkinsons Disease Campaign
The one complaint I have about 23andMe’s services is the monthly subscription fee. Paying even $5.00 a month is unacceptable, even for a great service, when it is accompanied with constant pressure to take research surveys to help the company determine the purpose of even more genetic markers. 23andMe appears to have the research to extrapolate the phenotypic expression of some genotypes, like my having curly hair, from just these surveys that no other genotyping service can offer. The site even features an advertisement with Muhammed Ali that they are using the surveys to hone in on the markers for Parkinson’s disease.
To be clear: I don’t mind the surveys and I don’t mind the subscription fee. I mind the combination of the two. I mind paying a company for access to their research while simultaneously contributing to their research to give them a proprietary edge over the competition. If 23andMe were a free service where you had to take one survey a month to maintain your access, I would think it the greatest thing in the world. The company has tremendous potential to harness information from its user community in the name of Citizen Science and reap fantastic profits from that data, but I’m not going to pay them to collect my data and then let them go off to make even more money from it.
Make access to the site conditional on buying a gene test from them and taking monthly surveys and I think 23andMe could be a wonderful thing for society.
Open-Source Genetics
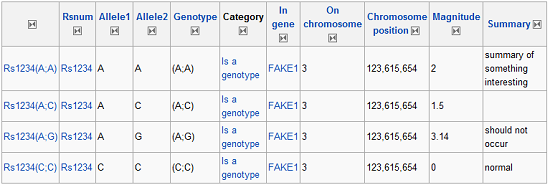
Because 23andMe lets you download your results, you are free to take them with you to other online services or even conduct your own research. That’s awesome and a bit daunting at the same time. Looking at a spreadsheet with nearly one-million SNPs on it doesn’t seem like the best place to start, and plugging SNPs Reference Cluster IDs (rs#) into google at random will only confuse you (despite the fact that you will get results from lots of databases). Probably the best source for randomly plugging in rs#’s is the National Center for Biotechnology Information’s (NCBI) Single-Nucleotide Polymorphism (SNP) database, where plugging in a random SNP will get you a wonderfully comprehensive block of information that looks something like this:
Partial View of NCBI Results
Incomprehensible to me, and I bet if someone were to explain it I would find that data fairly boring and irrelevant too.
So we’ll leave it to the many many many experts out there to figure out what’s interesting and what’s important. Luckily, a great, fairly down-to-earth resource I found online is SNPedia, a human genetics wiki. I’ll read about a trait or risk factor in the wiki and then run a “find” in my spreadsheet, locate the marker, and make a note beside it about what it means–but there’s an even better way.
Promethease

Promethease
SNPedia has a free software tool called Promethease that will take results from 23andMe or another service and run through your results to generate a custom report based on the information in the wiki. It takes about four hours to generate the report, but for a $2 Amazon payment, you can get the report in a few minutes (it took 412 seconds versus six hours for me running the report paid versus unpaid).
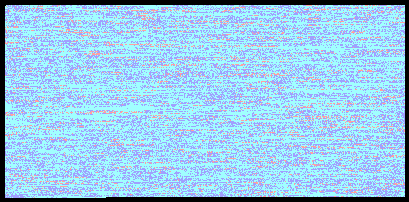
Ryan and Vicky Chromosome 1 Comparison
One of the cooler enhancements you get with the paid version of the report is the ability to compare your results to someone else. I ran my report with a comparison to Vicky’s results and got some experimental results back comparing our genomes. In the above image you can see our comparison for the first chromosome with light blue being a match (57%), dark blue being a halfmatch (37%), and the red being conflicts (6%).
A video tutorial of the features in the paid Promethease 0.1.99 version report shows some really neat additional comparison reports that did not show up in my report, such as seeing the probabilities of genotypes showing up in Vicky and my offspring and a Venn diagram of our genetic relatedness. I was unable to find out why, but I did find other ways of looking at my results in the report, such as the following screenshot (not that I’ve figured out what this means yet):
Promethease Visualization
The Promethease report gave me some additional tidbits about myself that 23andMe either didn’t provide or didn’t present as important such as my SNP for being “optimistic and empathetic” (rs53576(G;G), increased memory performance (rs17070145(C;T)), and better odds of living to 100 (rs2542052(C;C)). The information suffers the same issues of probability and environment as the 23andMe presentation, but the exercise demonstrates that it’s interesting and important to look at your genomic information in different ways.
DIY Genealogy with Personal Genomics
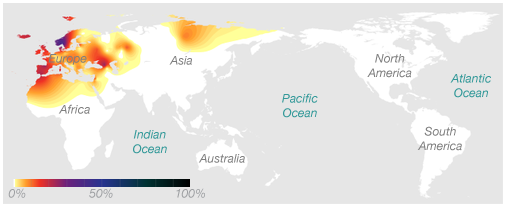
Ryan’s Maternal Haplogroup (h1)
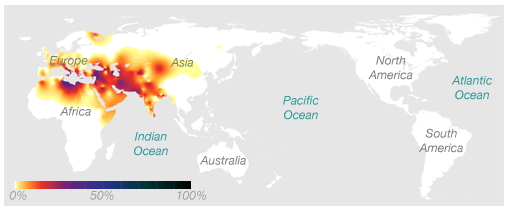
Ryan’s Paternal Haplogroup (J2)
Five years ago, I paid $100 to get my genealogical results from the Genographic Project, which took a DNA sample from my cheek and then showed me where in the course of human migrations my ancestors got off the caravan.
Using 23andMe’s Maternal and Paternal Line reading of our haplogroups, we can now save $100 and take our haplogroup information to National Geographic to find out for ourselves. Navigating to the Atlas of Human Journey, we can look up these haplogroups for more detailed information from their Genetic Markers list.
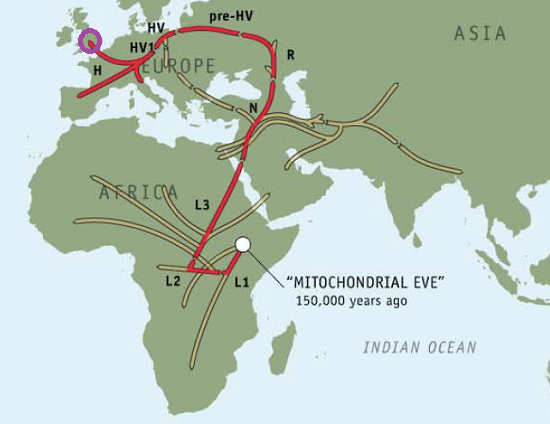
Genographic Project Maternal (H1)
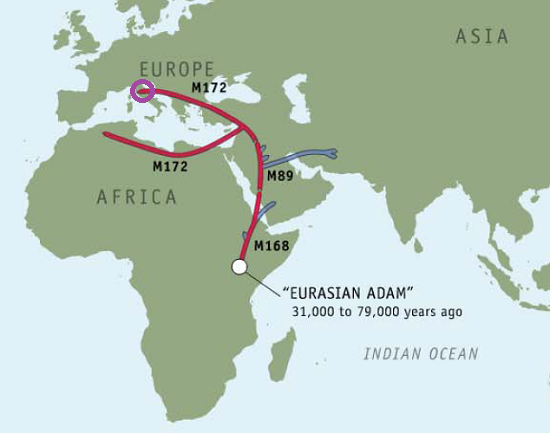
Genographic Project Paternal (J2)
International Society of Genetic Genealogy provides an index to Y-DNA SNPs that I, unfortunately, could not figure out how to use to divine my haplogroups from my raw data. Maybe in the future I’ll figure out how to get this information without going through 23andMe.
Going Public with My Genome
SNPedia has a long list of users who have shared their genomes with the online world that includes entrepreneur Esther Dyson, author Steven Pinker, biologist and entrepreneur Craig Venter, and Nobel-prize winner James D. Watson. PersonalGenomes.org also gathers genomic data from users who freely donate their results for citizen science.
When 23andMe had their genome test sale last year and I promoted it, I got a lot of feedback from friends who said they would never submit to such a test because they were afraid of the Government or Corporations getting ahold of that data. The Genetic Information Nondiscrimination Act of 2008 bars health insurance companies from denying you coverage for a genetic predisposition and employers from discriminating against you for what’s in your genotype. There are legitimate debates over whether the law goes too far or not far enough, but the intention is clear and based on the uncertainty of what’s revealed in a personal genomics test, the threat appears small.
But that does not mean I approach posting my genetic results lightly. SNPedia has a cautionary rs#, rs666, a fictional SNP that represents the worst possible genetic trait you could possibly imagine. What would it mean if future research revealed to the world that you had this genotype? Such a revelation would not only impact you, but your children as well.
I am posting my genetic data, raw and in reports, online for others to view (see below), but it is not something I do lightly. I believe information should be free, and I believe in the ideal of a world where I shouldn’t need to fear sharing this data.
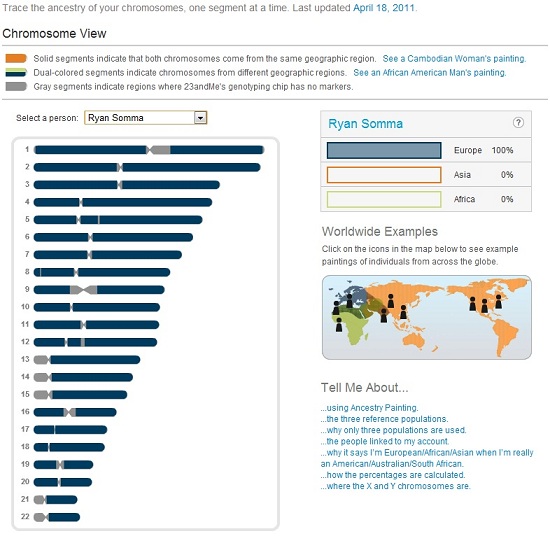
My 23andMe Ancestry Painting (100% European)
The Future of Healthcare with Personal Genomics
Erza Klein predicts personal genome sequencing spells the death of Health Insurance Companies:
As we sequence more genomes, mine more data, and conduct more studies, we’ll find a lot more of these connections. Eventually, genomic testing will be a powerful predictor of future illness. And it raises the potential that young people will get themselves tested and then purchase insurance based off the result. So those with a clean genomic result might go for a cheap catastrophic plan, while those with a high risk of developing pricey illnesses will opt for more comprehensive insurance.
The result would be, in insurance terms, an “adverse-selection death spiral,” as the healthy opt out of expensive insurance, the sick opt into it, and premiums spin out of control.
“For all of human history, humans have not had the readout of the software that makes them alive,” Larry Smarr, a member of the Complete Genomics scientific advisory board, told The New York Times. “Once you make the transition from a data poor to data rich environment, everything changes.”
The problem with this is, as I mentioned before, my results don’t predict my likelihood of getting a disease, they give me a better understanding of the odds to bet on, but if I have a 1.22-times chance of Type II Diabetes, there’s no way to customize my insurance plan to these odds. It’s all or nothing. Not only that, but would this even be legal? Congress has made it illegal for my insurance company to drop me for having an increased chance of manifesting one disease, so giving me a discount for having a lower propensity for another disease discriminates against those who have a higher propensity for it.
Better Living Through Personal Genomics
With states like New York and California having strict laws prohibiting or severely restricting genome tests or requiring the test results be explained by a doctor and the American Medical Association (AMA) lobbying the FDA to restrict access to our genomes, we have to wonder: What’s the harm?
My gut reaction is that doctors don’t like giving up their power. As I’ve discussed above, the data being provided only speaks to increased and decreased chances, and then we are talking about such tiny differences that it’s impossible to make an informed decision about what learned, especially in regards to cancelling a health insurance policy or forgoing doctor visits. Despite the best and worst-case prediction from both sides of the personal genomics debate, it would be foolish to do anything but work to improve your health after learning about your disease risks and genotypic traits.
Nature VS Nurture
My SNP Rs9834312 expresses as (G,G), which should make me an inch taller than other expressions, but I was a scrawny, malnourished nerd-boy in grade school, which stunted my growth to 5’9″ by the time I turned 20, about an inch shorter than the average American male. My lifestyle as a youth stunted my growth, but I don’t think knowing I had the genes for growing tall would have further encouraged that lifestyle. In fact, I think just the opposite would be true.
Knowledge of my genetic predispositions and education about how lifestyle choices impact phenotypic expression encourage people to take better care of themselves. The Navigenics website’s purpose statement hits the nail on the head that this information has the capacity “…to empower you with personal, confidential genetic insights to help motivate you to improve your health.” Knowing about our disease risks and other genetic shortcomings should inspire us to work harder for better health to improve our quality of life and odds of going further through it.
Appendix: DIY Genomic Sequencing for Programmers
This part of my article is technical, so feel free to skip it unless you are a computer programmer who wants to fill in the gaps in your genomic data.
SNPedia
SNPedia runs on MediaWiki, the same software running Wikipedia. Additionally, it runs Semantic mediawiki, meaning you can run semantic queries against it using the Ask Page:
| Query | Properties |
| [[Rsnum::Rs1234]] |
?Rsnum ?Allele1 ?Allele2 ?Genotype ?Category ?In gene ?On chromosome ?Chromosome position ?Magnitude ?Summary |
Which will produce results like so:
Semantic Results
Each of the nodes in this table has a “class” attribute with the property name, making it fairly easy to pluck the data from the table. You can then iterate through your results and query SNPedia with a query string like so, adjusting the RS# to match:
SNPedia also offer tips for getting bulk data, RDF Export (which I could not get to provide useful data), and MediaWiki API.
dbSNP
The NCBI also makes its data available for querying, but it’s more technical and complex to find useful information to align with your personal results. Using the list of SNP Database Fields and the Entrez ESearch Interface for query strings, you can get document summaries or SNP Properties in XML format.
dbSNP ER Diagram (Partial View)
Most intriguing is the fact that you can Dowload Entire dbSNP Database (179.81 GB) (MySQL), or query the database directly (see also database diagrams (PDF)).
My Personal Genomic Results
You can browse my Promethease report online here and I’ve got an XLSX spreadsheet with both my 23andMe and Promethease data here to download. Alternatively, you can download a tab-delimited copy of my 23andMe Results or my Annotated Promethease results.
Further Reading/Viewing
BBC The Ghost in your Genes documentary on epigenetics.
Esther Dyson Interview about her decision to Share her Genome.
Nature article on the Rise of Genome Bloggers.
Comments
3 responses to “Adventures in Personal Genomics”